Genetic analysis of both wild and hatchery-reared freshwater mussels is important to conservation programs interested in protecting and restoring mussel populations. Genetic analyses help biologists determine which populations are genetically similar and can be mixed to boost abundance and genetic variation within populations, and which populations are dissimilar and thus mixing should be avoided. Because the shell phenotypes of mussels are notoriously difficult to distinguish between some species, genetic data are also useful in delineating closely related or cryptic species.
We have conducted genetic analysis on numerous endangered mussel species, including the Oyster Mussel (Epioblasma capsaeformis), Duck River Dartersnapper (Epioblasma ahlstedti), Tan Riffleshell (Epioblasma florentina walkeri), Golden Riffleshell (Epioblasma florentina aureola), Northern Riffleshell (Epioblasma torulosa rangiana), Cumberlandian Combshell (Epioblasma brevidens), Fanshell (Cyprogenia stegaria), Dromedary Pearlymussel (Dromus dromas), Birdwing Pearlymussel (Lemiox rimosus), James Spinymussel (Pleurobema collina), Rough Pigtoe (Pleurobema plenum), the federal candidate species Slabside Pearlymussel (Lexingtonia dolabelloides), and even a few non-listed species such as the Wavy-rayed Lampmussel (Lampsilis fasciola). Genetic data are used to guide recovery efforts for these species and to understand aspects of their ecology, life history, and demography, such as providing estimates of migration rates among sub-populations and determining effective population size (Ne).
Our genetic analyses are conducted in collaboration with the department’s geneticist, Dr. Eric Hallerman. The genetics laboratory is a state-of-the-art facility located in the Integrated Life Sciences Building (ILSB) at the Corporate Research Center on Virginia Tech’s main campus in Blacksburg. The laboratory has ultra-low freezers for storing field collected samples and all the necessary equipment to conduct PCR amplification of DNA sequences and DNA microsatellites.
Shells (right valve shown) of the four species belonging to the Pleurbema cordatum complex: (A) Pleurbema plenum [Green River, Munfordville, Hart Co., KY], (B) Pleurbema cordatum [Green River, Munfordville, Hart Co., KY], (C) Pleurbema sintoxia [Ohio River, 5 miles SE of New Richmond, Campbell Co., KY] and (D) Pleurbema rubrum [Clinch River, Anderson Co., TN]. Shells and locality data are from the Ohio State University Museum.
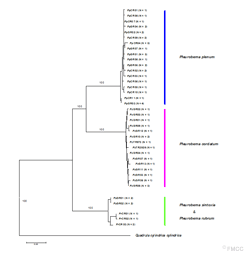
Phylogenetic relationships among the investigated Pleurobema species were inferred from the mitochondrial DNA region of ND1 (831 base pairs) using a Neighbor-Joing search algorithm. Numbers above the branches represent bootstrap support (1000 replicates) for the major clades. Numbers in parentheses at the end of each taxonomic name represent the number of observed haplotypes.